The basic question that I try to find an answer to is this:
Is it pure chance that methionine is the universal start amino acid in protein synthesis or is there a logical explanation why this amino acid and not any other (formyl-methionine doesn't count and glutamine or isoleucine are much less efficient at initiation of translation than methionine) is used.
Francis Crick wrote:
"Discussion of the actual amino acids used in the code may not be very profitable. Some less common amino acids, such as cysteine and histidine, would clearly seem to have an advantage because of their chemical reactivity; but whether, say, methionine could be justified in this way seems less obvious."
Crick FH. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367-79.
That may very well be true, but I think just opting for the "accident" solution is not a very satisfying answer and to have a logical explanation is better than not having any at all. Of course the structure of the initiator tRNA is the deciding factor in this, but it might have been conceivable that other initiator tRNAs might have been successful. If methionine is nearly always the first amino acid in a protein sequence, but often cleaved off with other N-terminal amino acids, then it might not be essential for the structure/function relation of the protein but still play an important role outside protein synthesis, which is where S-adenosylmethionine (AdoMet) comes in. S-adenosylmethionine is used in transmethylation (of DNA, rRNA, tRNA, mRNA-caps, proteins and lipids), transsulfuration, and aminopropylation (polyamine synthesis of spermidine and spermine). Could the fundamental processes of translation, transmethylation and polyamine synthesis be tethered or is it merely an interesting coincidence, a correlation in the absence of a "cause-and-effect" relation? How could one adress this question? How do you untangle a Gordian knot without using brute force that would wreck the whole system?
Tuesday, August 19, 2008
Friday, August 15, 2008
Mindgames with SAM-analogs
I keep on wondering what different S-adenosylmethionine analogs would do to the cellular metabolism and if such molecules might prove useful for investigating experimental leishmaniasis. Obviously, these molecules are likely to be toxic to the host as well as the parasite, but perhaps there might be a way to limit their exposure to macrophages, targeting amstigotes, and thereby reducing the expected undersired side effects to a minimum.
Compared to S-adenosylmethionine:
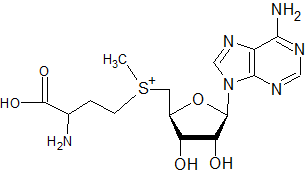
The following molecule would not be expected to act as a substrate in methylation (of DNA, rRNA, mRNA caps, tRNA, proteins, and phospholipids) or the transfer of propylamine, but not interfere with putrescine synthesis directly as sinefungin might:

This one could be able to catalyse the formation of spermidine and spermine, but lead to ethylation instead of methylation:

Whilst this molecule, if accepted by the respective enzymes as a substrate, could only be used for the transfer of propylamine but not for methylation/ethylation:

On the other hand, the following molecule would only act as a substrate for methylation and not for polyamine synthesis:

Whilst tubericidin is a SAM-analog that has been shown to affect L. donovani promastigotes but treatment has also led to tubericidin-resistant strains emerging:

In addition, attention should be paid to the stereoisomeric form of S-adenosylmethionine (AdoMet), the (Sc, Ss) isomer being the biologically active form, the (Sc, Rs) isomer being a potent inhibitor. (See review by Ronald Bentley).
Another idea that crossed my mind was whether methyl donor supplementation during pregnancy might render C57BL/6 or CBA mice more susceptible to leishmaniasis (similar to the results obtained for agouti mice regarding coat colour [Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003 Aug;23(15):5293-300.]).
Compared to S-adenosylmethionine:
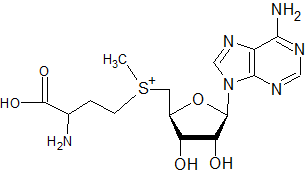
The following molecule would not be expected to act as a substrate in methylation (of DNA, rRNA, mRNA caps, tRNA, proteins, and phospholipids) or the transfer of propylamine, but not interfere with putrescine synthesis directly as sinefungin might:

This one could be able to catalyse the formation of spermidine and spermine, but lead to ethylation instead of methylation:

Whilst this molecule, if accepted by the respective enzymes as a substrate, could only be used for the transfer of propylamine but not for methylation/ethylation:

On the other hand, the following molecule would only act as a substrate for methylation and not for polyamine synthesis:

Whilst tubericidin is a SAM-analog that has been shown to affect L. donovani promastigotes but treatment has also led to tubericidin-resistant strains emerging:

In addition, attention should be paid to the stereoisomeric form of S-adenosylmethionine (AdoMet), the (Sc, Ss) isomer being the biologically active form, the (Sc, Rs) isomer being a potent inhibitor. (See review by Ronald Bentley).
Another idea that crossed my mind was whether methyl donor supplementation during pregnancy might render C57BL/6 or CBA mice more susceptible to leishmaniasis (similar to the results obtained for agouti mice regarding coat colour [Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003 Aug;23(15):5293-300.]).
Sunday, August 10, 2008
S-adenosylmethionine and sinefungin
There is always the possibility that I am only seeing what I want to see, which is in itself slightly unnerving and the reason why I repeatedly ask for feedback from anyone out there. Whether or not anyone who stumbles across these pages is actually involved in science and could potentially use these ideas is another matter. For the time being, all I can do is to write down my thoughts and hope that some day someone will know what to do with them.
I recently came across an old paper on antileishmanial properties of the S-adenosylmethionine-analog sinefungin:
Nolan LL. Molecular target of the antileishmanial action of sinefungin. Antimicrob Agents Chemother. 1987 Oct;31(10):1542-8.
We'll see if and how things may fit together, but sinefungin-mediated inhibition of polyamine synthesis or DNA methylation/ epigenetics might be possible mechanisms worth considering... In fact, sinefungin looks especially cool because it combines being a SAM/SAH-homologue as well as an ornithine derivative...
S-adenosylmethionine

(taken from "http://www.freepatentsonline.com/7048948-0-large.jpg")
Sinefungin

(taken from "http://www.sigmaaldrich.com/thumb/structureimages/59/s______s8559.gif")
Leishmania donovani was shown to be very sensitive to treatment with sinefungin, with even promastigotes being affected by the treatment. Which makes me start to wonder what might have happened to sinefungin in the pipeline to becoming a widely used antileishmanial drug? Is it possible to devise a delivery method for sinefungin which would target macrophages specifically, either by liposomes or linked to peptides that bind to macrophage receptors?
Bachrach U, Schnur LF, El-On J, Greenblatt CL, Pearlman E, Robert-Gero M, Lederer E. Inhibitory activity of sinefungin and SIBA (5'-deoxy-5'-S-isobutylthio-adenosine) on the growth of promastigotes and amastigotes of different species of Leishmania. FEBS Lett. 1980 Dec 1;121(2):287-91.
Phelouzat MA, Basselin M, Lawrence F, Robert-Gero M. Sinefungin shares AdoMet-uptake system to enter Leishmania donovani promastigotes. Biochem J. 1995 Jan 1;305 (Pt 1):133-7.
I recently came across an old paper on antileishmanial properties of the S-adenosylmethionine-analog sinefungin:
Nolan LL. Molecular target of the antileishmanial action of sinefungin. Antimicrob Agents Chemother. 1987 Oct;31(10):1542-8.
We'll see if and how things may fit together, but sinefungin-mediated inhibition of polyamine synthesis or DNA methylation/ epigenetics might be possible mechanisms worth considering... In fact, sinefungin looks especially cool because it combines being a SAM/SAH-homologue as well as an ornithine derivative...
S-adenosylmethionine

(taken from "http://www.freepatentsonline.com/7048948-0-large.jpg")
Sinefungin

(taken from "http://www.sigmaaldrich.com/thumb/structureimages/59/s______s8559.gif")
Leishmania donovani was shown to be very sensitive to treatment with sinefungin, with even promastigotes being affected by the treatment. Which makes me start to wonder what might have happened to sinefungin in the pipeline to becoming a widely used antileishmanial drug? Is it possible to devise a delivery method for sinefungin which would target macrophages specifically, either by liposomes or linked to peptides that bind to macrophage receptors?
Bachrach U, Schnur LF, El-On J, Greenblatt CL, Pearlman E, Robert-Gero M, Lederer E. Inhibitory activity of sinefungin and SIBA (5'-deoxy-5'-S-isobutylthio-adenosine) on the growth of promastigotes and amastigotes of different species of Leishmania. FEBS Lett. 1980 Dec 1;121(2):287-91.
Phelouzat MA, Basselin M, Lawrence F, Robert-Gero M. Sinefungin shares AdoMet-uptake system to enter Leishmania donovani promastigotes. Biochem J. 1995 Jan 1;305 (Pt 1):133-7.
Thursday, August 07, 2008
This is a tRNA's world, but it would nothing, not one little thing, without an amino acid or a peptide...
With the print version of my contribution to the Journal of Theoretical Biology made available today, here's a question that has been on my mind for quite some time.
I keep on wondering, in what way the basic components of the genetic code may have evolved in the absence of any translational machinery. In other words, is it possible that precursors to today's tRNAs simply bound to amino acids thus enabling them to accumulate in higher concentrations than would have been possible in the absence of such interactions? tRNA-like molecules that bound to hydrophobic amino acids may have been able to interact with lipids at the liquid/air or at the liquid/solid interphase. With an accumulation of those tRNA-like molecules other tRNA-like molecules that bound to hydrophilic amino acids may have in turn formed aggregates with the hydrophobic amino acid-binding tRNAs via the codon-equivalent regions, leading to the basic dichotomy between N-U-N and N'-A-N' codons and to a microenvironment with favourable conditions for interactions between different species of nucleic and amino acids. The hydrophilic amino acids would have to balance the charge inequalities leading to basic and acidic amino acids landing close codon proximity. The next step would be that RNA molecules with catalytic activity bound these adaptor RNAs at strategic positions and the attached amino acids started to be a part if the catalytic process. Finally, the translational machinery would have evolved and the code would have continued to change with it. Some RNA-amino acid pairings would have been thrown out of the race and new ones joined the process at this stage. A drive to reduce codon ambiguity and to minimize errors in translation are likely to be two important factors in the evolution of the code, but some initial rules laid down by the pre-translation system were already in place before the era of large-scale protein synthesis.
I am thankful for any comments you might care to send my way.
If you are interested in more literature on the genetic code, I can recommend:
Nirenberg, MW, Matthaei, JH, Jones, OW. An intermediate in the biosynthesis of polyphenylalanine directed by synthetic template RNA. Proc Natl Acad Sci U S A. 1962 Jan 15;48:104-9.
Woese, CR. On the evolution of the genetic code. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1546-52.
Crick, FH. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367-79.
Orgel, LE. Evolution of the genetic apparatus. J Mol Biol. 1968 Dec;38(3):381-93.
Wong, JT. A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A. 1975 May;72(5):1909-12.
Taylor, FJ, Coates, D. The code within the codons. Biosystems. 1989;22(3):177-87.
Szathmáry, E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends in Genetics. 1999 June;15(6): 223-9.
Massey, SE. A sequential "2-1-3" model of genetic code evolution that explains codon constraints. J Mol Evol. 2006 Jun;62(6):809-10. Epub 2006 Apr 11.
I keep on wondering, in what way the basic components of the genetic code may have evolved in the absence of any translational machinery. In other words, is it possible that precursors to today's tRNAs simply bound to amino acids thus enabling them to accumulate in higher concentrations than would have been possible in the absence of such interactions? tRNA-like molecules that bound to hydrophobic amino acids may have been able to interact with lipids at the liquid/air or at the liquid/solid interphase. With an accumulation of those tRNA-like molecules other tRNA-like molecules that bound to hydrophilic amino acids may have in turn formed aggregates with the hydrophobic amino acid-binding tRNAs via the codon-equivalent regions, leading to the basic dichotomy between N-U-N and N'-A-N' codons and to a microenvironment with favourable conditions for interactions between different species of nucleic and amino acids. The hydrophilic amino acids would have to balance the charge inequalities leading to basic and acidic amino acids landing close codon proximity. The next step would be that RNA molecules with catalytic activity bound these adaptor RNAs at strategic positions and the attached amino acids started to be a part if the catalytic process. Finally, the translational machinery would have evolved and the code would have continued to change with it. Some RNA-amino acid pairings would have been thrown out of the race and new ones joined the process at this stage. A drive to reduce codon ambiguity and to minimize errors in translation are likely to be two important factors in the evolution of the code, but some initial rules laid down by the pre-translation system were already in place before the era of large-scale protein synthesis.
I am thankful for any comments you might care to send my way.
If you are interested in more literature on the genetic code, I can recommend:
Nirenberg, MW, Matthaei, JH, Jones, OW. An intermediate in the biosynthesis of polyphenylalanine directed by synthetic template RNA. Proc Natl Acad Sci U S A. 1962 Jan 15;48:104-9.
Woese, CR. On the evolution of the genetic code. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1546-52.
Crick, FH. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367-79.
Orgel, LE. Evolution of the genetic apparatus. J Mol Biol. 1968 Dec;38(3):381-93.
Wong, JT. A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A. 1975 May;72(5):1909-12.
Taylor, FJ, Coates, D. The code within the codons. Biosystems. 1989;22(3):177-87.
Szathmáry, E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends in Genetics. 1999 June;15(6): 223-9.
Massey, SE. A sequential "2-1-3" model of genetic code evolution that explains codon constraints. J Mol Evol. 2006 Jun;62(6):809-10. Epub 2006 Apr 11.
Labels:
evolution,
Genetic code,
translation
Thursday, July 31, 2008
SOCS3 expression in classically activated rat macrophages impairs arginase expression
Well, after nearly dismissing my ideas on SOCS and leishmaniasis in my last post, I came across this interesting paper by Liu and colleagues that rekindled the old flame a bit. Published in the Journal of Immunology in May 2008, they show that SOCS3 knockdown by RNA interference in classically activated rat macrophages coincided with an upregulation of alternative activation makers such as arginase.
Liu Y, Stewart KN, Bishop E, Marek CJ, Kluth DC, Rees AJ, Wilson HM. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J Immunol. 2008 May 1;180(9):6270-8.

This brings me back to my question: would SOCS3 shut down STAT3 (IL-6-induced arginase) and IRS-2 signalling and SOCS5 shut down STAT6 (IL-4-induced arginase) signalling so that arginase activity would drop to levels below those able to support parasite proliferation? What role does S-adenosylmethionine play in all this - control of SOCS expression via DNA methylation versus polyamine synthesis of spermine and spermidine? Untangling all this experimentally will no doubt be a bit tricky, because the effects of SOCS on macrophages should be kept separate from effects of SOCS on other cell types.
I would suggest to start in vitro and to compare the effects of RNAi mediated by siRNA against SOCS1, SOCS3, SOCS5, DNA (cytosine-5-)-methyltransferase 1, DNA (cytosine-5-)-methyltransferase 3 alpha, DNA (cytosine-5-)-methyltransferase 3 beta, S-adenosylmethionine synthetase, ornithine decarboxylase, S-adenosylmethionine decarboxylase, spermidine synthase and spermine synthase in respect to parasite development. An interesting side project would be to find out whether or not culturing macrophages in the presence of increased S-adenosylmethionine concentrations leads to higher parasite proliferation. The in vivo experiments that spring to mind are inducible macrophage-specific expression of transgenes: either SOCS-specific shRNA in resistant mice or SOCS in susceptible mice. Whilst it would be preferable if the gene expression was not leaky, I think my wish list is getting slightly too demanding, but if anyone out there thinks that some of these ideas are useful and has the means to set up the necessary experiments please feel welcome to do so. I believe that the sooner the scientific community can come up with new affordable drugs against this neglected tropical disease the better. I would argue the best way to achieve this is through cooperation rather than competition. As always, feedback is greatly appreciated.
Liu Y, Stewart KN, Bishop E, Marek CJ, Kluth DC, Rees AJ, Wilson HM. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J Immunol. 2008 May 1;180(9):6270-8.

This brings me back to my question: would SOCS3 shut down STAT3 (IL-6-induced arginase) and IRS-2 signalling and SOCS5 shut down STAT6 (IL-4-induced arginase) signalling so that arginase activity would drop to levels below those able to support parasite proliferation? What role does S-adenosylmethionine play in all this - control of SOCS expression via DNA methylation versus polyamine synthesis of spermine and spermidine? Untangling all this experimentally will no doubt be a bit tricky, because the effects of SOCS on macrophages should be kept separate from effects of SOCS on other cell types.
I would suggest to start in vitro and to compare the effects of RNAi mediated by siRNA against SOCS1, SOCS3, SOCS5, DNA (cytosine-5-)-methyltransferase 1, DNA (cytosine-5-)-methyltransferase 3 alpha, DNA (cytosine-5-)-methyltransferase 3 beta, S-adenosylmethionine synthetase, ornithine decarboxylase, S-adenosylmethionine decarboxylase, spermidine synthase and spermine synthase in respect to parasite development. An interesting side project would be to find out whether or not culturing macrophages in the presence of increased S-adenosylmethionine concentrations leads to higher parasite proliferation. The in vivo experiments that spring to mind are inducible macrophage-specific expression of transgenes: either SOCS-specific shRNA in resistant mice or SOCS in susceptible mice. Whilst it would be preferable if the gene expression was not leaky, I think my wish list is getting slightly too demanding, but if anyone out there thinks that some of these ideas are useful and has the means to set up the necessary experiments please feel welcome to do so. I believe that the sooner the scientific community can come up with new affordable drugs against this neglected tropical disease the better. I would argue the best way to achieve this is through cooperation rather than competition. As always, feedback is greatly appreciated.
Labels:
arginase,
immunology,
leishmania,
SAM,
SOCS
Monday, May 26, 2008
Arginase and ageing - rethinking SAM
Age-Related Alteration of Arginase Activity Impacts on Severity of Leishmaniasis
is a very exciting paper, so convincing in fact that it makes me question whether my hypothesis on SAM is of any use at all to explain susceptibility to leishmaniasis. Arginase activity has been shown time and time again to be crucial to parasite proliferation, but I always wondered whether there was some proverbial elephant standing in the room that no-one mentioned. The SAM story was meant to complement and expand the arginase part and I still believe that with its dual role in polyamine synthesis and regulation of gene expression amongst others, SAM might still be shown to play a role in this experimental system.
At first, I thought that by changes in DNA methylation SOCS expression would be altered affecting cell signalling and arginase activity in host macrophages. This may still be the case, but maybe it would be better to focus on the polyamine synthesis aspect at first. If antimony treatment should deplete SAM levels this could be investigated by incubating macrophage cultures with antimony and measure polyamine levels by thin layer chromatography as carried out by Modolell and colleagues.

is a very exciting paper, so convincing in fact that it makes me question whether my hypothesis on SAM is of any use at all to explain susceptibility to leishmaniasis. Arginase activity has been shown time and time again to be crucial to parasite proliferation, but I always wondered whether there was some proverbial elephant standing in the room that no-one mentioned. The SAM story was meant to complement and expand the arginase part and I still believe that with its dual role in polyamine synthesis and regulation of gene expression amongst others, SAM might still be shown to play a role in this experimental system.
At first, I thought that by changes in DNA methylation SOCS expression would be altered affecting cell signalling and arginase activity in host macrophages. This may still be the case, but maybe it would be better to focus on the polyamine synthesis aspect at first. If antimony treatment should deplete SAM levels this could be investigated by incubating macrophage cultures with antimony and measure polyamine levels by thin layer chromatography as carried out by Modolell and colleagues.

Labels:
arginase,
immunology,
leishmania,
SAM
Friday, May 09, 2008
Letting go - now back to Leishmania
Letting go is hard to do, especially when you have fought tooth and nail to get your point across and went through some crazy times. I recently attended the "God Be in My Head" event at the Dana Centre, where a panel chaired by Colin Blakemore discussed the possibility of so-called "God spots" in the brain, and although epilepsy and schizzophrenia were mentioned as possible links to abstract thinking, creativity and spiritual experience, I was surprised to observe that no-one at the event mentioned mania, which seems to me to lie at the intersection between the two other states of mind. One of the best suggestions of the evening was the idea to analyse the brain activity of problem-solving scientists using functional MRI and to try to compare the results to the brain scans of nuns praying or monks meditating. I like it even though I am getting a bit tired of functional MRI papers, but somehow I don't see that experiment being carried out any time soon, the eureka experience and phenomena such as calculation-induced halucinations are science's dirty little secrets and I doubt whether scientists really are prepared to address these issues.
At any rate, I'm digressing, whilst it is fun to rejoin the scientific debate and to watch my representation of the genetic code climb the "google charts", I have to let go of certain ideas and redirect my focus on my PhD. Consequently, I find myself thinking more and more about Leishmania these days. Given that S-adenosyl methionine (SAM) is involved in a number of different cellular processes possibly affecting the replication of intracellular parasites, how would it be possible to differentiate the effects of the individual processes? The ones I am particularly interested in are polyamine synthesis and DNA methylation. Would it be enough to add radiolabelled SAM to infected cells and to try to see where the marked methyl group might end up? Is there radiolabelled SAM available in which the non-methyl carbon atoms are 14C, making the substance useful in tracking polyamines? Whilst I would try to focus on in vitro experiments at first keeping things as simple as possible, I also wonder what in vivo experiments might look like. What effect might adding or depleting SAM have in vivo? What about methylthioadenosine (MTA), a product of the SAM catabolism that arises after the polyamine synthesis step? How could the effect of SAM derivatives be untangled from possible changes to cytokine production?
Too many questions for me to answer...
At any rate, I'm digressing, whilst it is fun to rejoin the scientific debate and to watch my representation of the genetic code climb the "google charts", I have to let go of certain ideas and redirect my focus on my PhD. Consequently, I find myself thinking more and more about Leishmania these days. Given that S-adenosyl methionine (SAM) is involved in a number of different cellular processes possibly affecting the replication of intracellular parasites, how would it be possible to differentiate the effects of the individual processes? The ones I am particularly interested in are polyamine synthesis and DNA methylation. Would it be enough to add radiolabelled SAM to infected cells and to try to see where the marked methyl group might end up? Is there radiolabelled SAM available in which the non-methyl carbon atoms are 14C, making the substance useful in tracking polyamines? Whilst I would try to focus on in vitro experiments at first keeping things as simple as possible, I also wonder what in vivo experiments might look like. What effect might adding or depleting SAM have in vivo? What about methylthioadenosine (MTA), a product of the SAM catabolism that arises after the polyamine synthesis step? How could the effect of SAM derivatives be untangled from possible changes to cytokine production?
Too many questions for me to answer...
Friday, May 02, 2008
Nearly there...
Finally, the "Journal of Theoretical Biology" accepted my manuscript for publication as a letter to the editor. The unedited version of the article can be found ahead of print online at
http://dx.doi.org/10.1016/j.jtbi.2008.04.028.
What a relief. I know it's just one little paper and the impact factor of the journal could be higher, but for some reason this paper is incredibly important to me. I don't know if I have made a huge fool of myself or not, but it's too late anyway. I tried to keep the article as short as possible and did not include any acknowledgements, but of course I would not have been able to persevere in this ridiculous struggle had I not had the support of some incredible people along the way. I don't know whether mRNA-tRNA interaction occurs in a 2-1-2-3 way, but I find the sheer oddness of a language that starts with the middle instead of the beginning of an information unit exciting and cannot get the picture of tRNA-precursor molecules forming aggregates with the second codon base acting as an anchor out of my head. Imagine the message
"hteatsiksontosmcuhotseewahtonoenhsayteseenubtottihnwkhtanoonheaysetthuogthaobutthtawihcehvreyobdsyese."
being translated as "thetaskisnotsomuchtoseewhatnoonehasyetseenbuttothinkwhatnoonehasyetthoughtaboutthatwhicheverybodysees." and finally for the information to "fold" into the beautiful quote: "The task is not so much to see what no one has yet seen but to think what no one has yet thought, about that which everybody sees."
Of course if the interaction between mRNA and tRNA occurred in a 1-2-3-way, the importance of the second base might be explained by a longer interval spent reading the base, so the order would roughly be 1-2-2-3. If the binding of codon and anticodon happened in such a way that all three bases bound simultaneously, then the base in the middle might spend the longest time bound to its cognate because the bases 5' and 3' of it would act as a sort of buffer (like velcro, the middle bit usually the hardest to get off first).
With possible closure on the rearrangement of the genetic code in sight, I wonder if somebody else might pick up on the methionine story. Perhaps one group might try to engineer an organism that uses an initiator-tRNA charged with isoleucine, valine, threonine or homocysteine instead of methionine and report on what the resulting phenotype might be...
But all of this has merely been a side project, the biggest reward for me would be if my initial hypothesis about the involvement of S-adenosylmethionine in determining resistance or susceptibility in experimental leishmaniasis, might prove to be useful.
http://dx.doi.org/10.1016/j.jtbi.2008.04.028.
What a relief. I know it's just one little paper and the impact factor of the journal could be higher, but for some reason this paper is incredibly important to me. I don't know if I have made a huge fool of myself or not, but it's too late anyway. I tried to keep the article as short as possible and did not include any acknowledgements, but of course I would not have been able to persevere in this ridiculous struggle had I not had the support of some incredible people along the way. I don't know whether mRNA-tRNA interaction occurs in a 2-1-2-3 way, but I find the sheer oddness of a language that starts with the middle instead of the beginning of an information unit exciting and cannot get the picture of tRNA-precursor molecules forming aggregates with the second codon base acting as an anchor out of my head. Imagine the message
"hteatsiksontosmcuhotseewahtonoenhsayteseenubtottihnwkhtanoonheaysetthuogthaobutthtawihcehvreyobdsyese."
being translated as "thetaskisnotsomuchtoseewhatnoonehasyetseenbuttothinkwhatnoonehasyetthoughtaboutthatwhicheverybodysees." and finally for the information to "fold" into the beautiful quote: "The task is not so much to see what no one has yet seen but to think what no one has yet thought, about that which everybody sees."
Of course if the interaction between mRNA and tRNA occurred in a 1-2-3-way, the importance of the second base might be explained by a longer interval spent reading the base, so the order would roughly be 1-2-2-3. If the binding of codon and anticodon happened in such a way that all three bases bound simultaneously, then the base in the middle might spend the longest time bound to its cognate because the bases 5' and 3' of it would act as a sort of buffer (like velcro, the middle bit usually the hardest to get off first).
With possible closure on the rearrangement of the genetic code in sight, I wonder if somebody else might pick up on the methionine story. Perhaps one group might try to engineer an organism that uses an initiator-tRNA charged with isoleucine, valine, threonine or homocysteine instead of methionine and report on what the resulting phenotype might be...
But all of this has merely been a side project, the biggest reward for me would be if my initial hypothesis about the involvement of S-adenosylmethionine in determining resistance or susceptibility in experimental leishmaniasis, might prove to be useful.
Sunday, April 27, 2008
Early nerdcore
Over at the excellent www.biology-blog.com I came across this wonderful video mixing free love and protein synthesis (trippy translation). I want more of this stuff: "The Knife" in charge of TLR signalling, "Roisin Murphy" could sing about MHC class I presentation whilst "Hot Chip" serenade MHC class II, cathepsins and lysosomes and "Portishead" would present apoptosis like no-one has ever done before...
Science classes would never be the same again...
Here's a brilliant clip I came across in 2009: Carl Sagan rocks!
Labels:
Science communication,
translation
Subscribe to:
Posts (Atom)