The basic question that I try to find an answer to is this:
Is it pure chance that methionine is the universal start amino acid in protein synthesis or is there a logical explanation why this amino acid and not any other (formyl-methionine doesn't count and glutamine or isoleucine are much less efficient at initiation of translation than methionine) is used.
Francis Crick wrote:
"Discussion of the actual amino acids used in the code may not be very profitable. Some less common amino acids, such as cysteine and histidine, would clearly seem to have an advantage because of their chemical reactivity; but whether, say, methionine could be justified in this way seems less obvious."
Crick FH. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367-79.
That may very well be true, but I think just opting for the "accident" solution is not a very satisfying answer and to have a logical explanation is better than not having any at all. Of course the structure of the initiator tRNA is the deciding factor in this, but it might have been conceivable that other initiator tRNAs might have been successful. If methionine is nearly always the first amino acid in a protein sequence, but often cleaved off with other N-terminal amino acids, then it might not be essential for the structure/function relation of the protein but still play an important role outside protein synthesis, which is where S-adenosylmethionine (AdoMet) comes in. S-adenosylmethionine is used in transmethylation (of DNA, rRNA, tRNA, mRNA-caps, proteins and lipids), transsulfuration, and aminopropylation (polyamine synthesis of spermidine and spermine). Could the fundamental processes of translation, transmethylation and polyamine synthesis be tethered or is it merely an interesting coincidence, a correlation in the absence of a "cause-and-effect" relation? How could one adress this question? How do you untangle a Gordian knot without using brute force that would wreck the whole system?
Tuesday, August 19, 2008
Friday, August 15, 2008
Mindgames with SAM-analogs
I keep on wondering what different S-adenosylmethionine analogs would do to the cellular metabolism and if such molecules might prove useful for investigating experimental leishmaniasis. Obviously, these molecules are likely to be toxic to the host as well as the parasite, but perhaps there might be a way to limit their exposure to macrophages, targeting amstigotes, and thereby reducing the expected undersired side effects to a minimum.
Compared to S-adenosylmethionine:
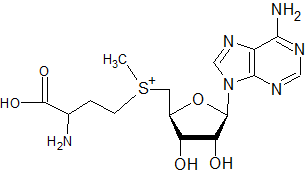
The following molecule would not be expected to act as a substrate in methylation (of DNA, rRNA, mRNA caps, tRNA, proteins, and phospholipids) or the transfer of propylamine, but not interfere with putrescine synthesis directly as sinefungin might:

This one could be able to catalyse the formation of spermidine and spermine, but lead to ethylation instead of methylation:

Whilst this molecule, if accepted by the respective enzymes as a substrate, could only be used for the transfer of propylamine but not for methylation/ethylation:

On the other hand, the following molecule would only act as a substrate for methylation and not for polyamine synthesis:

Whilst tubericidin is a SAM-analog that has been shown to affect L. donovani promastigotes but treatment has also led to tubericidin-resistant strains emerging:

In addition, attention should be paid to the stereoisomeric form of S-adenosylmethionine (AdoMet), the (Sc, Ss) isomer being the biologically active form, the (Sc, Rs) isomer being a potent inhibitor. (See review by Ronald Bentley).
Another idea that crossed my mind was whether methyl donor supplementation during pregnancy might render C57BL/6 or CBA mice more susceptible to leishmaniasis (similar to the results obtained for agouti mice regarding coat colour [Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003 Aug;23(15):5293-300.]).
Compared to S-adenosylmethionine:
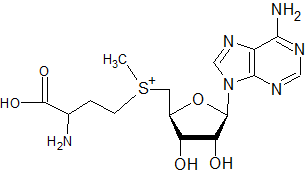
The following molecule would not be expected to act as a substrate in methylation (of DNA, rRNA, mRNA caps, tRNA, proteins, and phospholipids) or the transfer of propylamine, but not interfere with putrescine synthesis directly as sinefungin might:

This one could be able to catalyse the formation of spermidine and spermine, but lead to ethylation instead of methylation:

Whilst this molecule, if accepted by the respective enzymes as a substrate, could only be used for the transfer of propylamine but not for methylation/ethylation:

On the other hand, the following molecule would only act as a substrate for methylation and not for polyamine synthesis:

Whilst tubericidin is a SAM-analog that has been shown to affect L. donovani promastigotes but treatment has also led to tubericidin-resistant strains emerging:

In addition, attention should be paid to the stereoisomeric form of S-adenosylmethionine (AdoMet), the (Sc, Ss) isomer being the biologically active form, the (Sc, Rs) isomer being a potent inhibitor. (See review by Ronald Bentley).
Another idea that crossed my mind was whether methyl donor supplementation during pregnancy might render C57BL/6 or CBA mice more susceptible to leishmaniasis (similar to the results obtained for agouti mice regarding coat colour [Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003 Aug;23(15):5293-300.]).
Sunday, August 10, 2008
S-adenosylmethionine and sinefungin
There is always the possibility that I am only seeing what I want to see, which is in itself slightly unnerving and the reason why I repeatedly ask for feedback from anyone out there. Whether or not anyone who stumbles across these pages is actually involved in science and could potentially use these ideas is another matter. For the time being, all I can do is to write down my thoughts and hope that some day someone will know what to do with them.
I recently came across an old paper on antileishmanial properties of the S-adenosylmethionine-analog sinefungin:
Nolan LL. Molecular target of the antileishmanial action of sinefungin. Antimicrob Agents Chemother. 1987 Oct;31(10):1542-8.
We'll see if and how things may fit together, but sinefungin-mediated inhibition of polyamine synthesis or DNA methylation/ epigenetics might be possible mechanisms worth considering... In fact, sinefungin looks especially cool because it combines being a SAM/SAH-homologue as well as an ornithine derivative...
S-adenosylmethionine

(taken from "http://www.freepatentsonline.com/7048948-0-large.jpg")
Sinefungin

(taken from "http://www.sigmaaldrich.com/thumb/structureimages/59/s______s8559.gif")
Leishmania donovani was shown to be very sensitive to treatment with sinefungin, with even promastigotes being affected by the treatment. Which makes me start to wonder what might have happened to sinefungin in the pipeline to becoming a widely used antileishmanial drug? Is it possible to devise a delivery method for sinefungin which would target macrophages specifically, either by liposomes or linked to peptides that bind to macrophage receptors?
Bachrach U, Schnur LF, El-On J, Greenblatt CL, Pearlman E, Robert-Gero M, Lederer E. Inhibitory activity of sinefungin and SIBA (5'-deoxy-5'-S-isobutylthio-adenosine) on the growth of promastigotes and amastigotes of different species of Leishmania. FEBS Lett. 1980 Dec 1;121(2):287-91.
Phelouzat MA, Basselin M, Lawrence F, Robert-Gero M. Sinefungin shares AdoMet-uptake system to enter Leishmania donovani promastigotes. Biochem J. 1995 Jan 1;305 (Pt 1):133-7.
I recently came across an old paper on antileishmanial properties of the S-adenosylmethionine-analog sinefungin:
Nolan LL. Molecular target of the antileishmanial action of sinefungin. Antimicrob Agents Chemother. 1987 Oct;31(10):1542-8.
We'll see if and how things may fit together, but sinefungin-mediated inhibition of polyamine synthesis or DNA methylation/ epigenetics might be possible mechanisms worth considering... In fact, sinefungin looks especially cool because it combines being a SAM/SAH-homologue as well as an ornithine derivative...
S-adenosylmethionine

(taken from "http://www.freepatentsonline.com/7048948-0-large.jpg")
Sinefungin

(taken from "http://www.sigmaaldrich.com/thumb/structureimages/59/s______s8559.gif")
Leishmania donovani was shown to be very sensitive to treatment with sinefungin, with even promastigotes being affected by the treatment. Which makes me start to wonder what might have happened to sinefungin in the pipeline to becoming a widely used antileishmanial drug? Is it possible to devise a delivery method for sinefungin which would target macrophages specifically, either by liposomes or linked to peptides that bind to macrophage receptors?
Bachrach U, Schnur LF, El-On J, Greenblatt CL, Pearlman E, Robert-Gero M, Lederer E. Inhibitory activity of sinefungin and SIBA (5'-deoxy-5'-S-isobutylthio-adenosine) on the growth of promastigotes and amastigotes of different species of Leishmania. FEBS Lett. 1980 Dec 1;121(2):287-91.
Phelouzat MA, Basselin M, Lawrence F, Robert-Gero M. Sinefungin shares AdoMet-uptake system to enter Leishmania donovani promastigotes. Biochem J. 1995 Jan 1;305 (Pt 1):133-7.
Thursday, August 07, 2008
This is a tRNA's world, but it would nothing, not one little thing, without an amino acid or a peptide...
With the print version of my contribution to the Journal of Theoretical Biology made available today, here's a question that has been on my mind for quite some time.
I keep on wondering, in what way the basic components of the genetic code may have evolved in the absence of any translational machinery. In other words, is it possible that precursors to today's tRNAs simply bound to amino acids thus enabling them to accumulate in higher concentrations than would have been possible in the absence of such interactions? tRNA-like molecules that bound to hydrophobic amino acids may have been able to interact with lipids at the liquid/air or at the liquid/solid interphase. With an accumulation of those tRNA-like molecules other tRNA-like molecules that bound to hydrophilic amino acids may have in turn formed aggregates with the hydrophobic amino acid-binding tRNAs via the codon-equivalent regions, leading to the basic dichotomy between N-U-N and N'-A-N' codons and to a microenvironment with favourable conditions for interactions between different species of nucleic and amino acids. The hydrophilic amino acids would have to balance the charge inequalities leading to basic and acidic amino acids landing close codon proximity. The next step would be that RNA molecules with catalytic activity bound these adaptor RNAs at strategic positions and the attached amino acids started to be a part if the catalytic process. Finally, the translational machinery would have evolved and the code would have continued to change with it. Some RNA-amino acid pairings would have been thrown out of the race and new ones joined the process at this stage. A drive to reduce codon ambiguity and to minimize errors in translation are likely to be two important factors in the evolution of the code, but some initial rules laid down by the pre-translation system were already in place before the era of large-scale protein synthesis.
I am thankful for any comments you might care to send my way.
If you are interested in more literature on the genetic code, I can recommend:
Nirenberg, MW, Matthaei, JH, Jones, OW. An intermediate in the biosynthesis of polyphenylalanine directed by synthetic template RNA. Proc Natl Acad Sci U S A. 1962 Jan 15;48:104-9.
Woese, CR. On the evolution of the genetic code. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1546-52.
Crick, FH. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367-79.
Orgel, LE. Evolution of the genetic apparatus. J Mol Biol. 1968 Dec;38(3):381-93.
Wong, JT. A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A. 1975 May;72(5):1909-12.
Taylor, FJ, Coates, D. The code within the codons. Biosystems. 1989;22(3):177-87.
Szathmáry, E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends in Genetics. 1999 June;15(6): 223-9.
Massey, SE. A sequential "2-1-3" model of genetic code evolution that explains codon constraints. J Mol Evol. 2006 Jun;62(6):809-10. Epub 2006 Apr 11.
I keep on wondering, in what way the basic components of the genetic code may have evolved in the absence of any translational machinery. In other words, is it possible that precursors to today's tRNAs simply bound to amino acids thus enabling them to accumulate in higher concentrations than would have been possible in the absence of such interactions? tRNA-like molecules that bound to hydrophobic amino acids may have been able to interact with lipids at the liquid/air or at the liquid/solid interphase. With an accumulation of those tRNA-like molecules other tRNA-like molecules that bound to hydrophilic amino acids may have in turn formed aggregates with the hydrophobic amino acid-binding tRNAs via the codon-equivalent regions, leading to the basic dichotomy between N-U-N and N'-A-N' codons and to a microenvironment with favourable conditions for interactions between different species of nucleic and amino acids. The hydrophilic amino acids would have to balance the charge inequalities leading to basic and acidic amino acids landing close codon proximity. The next step would be that RNA molecules with catalytic activity bound these adaptor RNAs at strategic positions and the attached amino acids started to be a part if the catalytic process. Finally, the translational machinery would have evolved and the code would have continued to change with it. Some RNA-amino acid pairings would have been thrown out of the race and new ones joined the process at this stage. A drive to reduce codon ambiguity and to minimize errors in translation are likely to be two important factors in the evolution of the code, but some initial rules laid down by the pre-translation system were already in place before the era of large-scale protein synthesis.
I am thankful for any comments you might care to send my way.
If you are interested in more literature on the genetic code, I can recommend:
Nirenberg, MW, Matthaei, JH, Jones, OW. An intermediate in the biosynthesis of polyphenylalanine directed by synthetic template RNA. Proc Natl Acad Sci U S A. 1962 Jan 15;48:104-9.
Woese, CR. On the evolution of the genetic code. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1546-52.
Crick, FH. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367-79.
Orgel, LE. Evolution of the genetic apparatus. J Mol Biol. 1968 Dec;38(3):381-93.
Wong, JT. A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A. 1975 May;72(5):1909-12.
Taylor, FJ, Coates, D. The code within the codons. Biosystems. 1989;22(3):177-87.
Szathmáry, E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends in Genetics. 1999 June;15(6): 223-9.
Massey, SE. A sequential "2-1-3" model of genetic code evolution that explains codon constraints. J Mol Evol. 2006 Jun;62(6):809-10. Epub 2006 Apr 11.
Labels:
evolution,
Genetic code,
translation
Subscribe to:
Posts (Atom)